Dataset Structure
Default Dataset Setup¤
The AUCMEDI AutoML expects a fixed dataset structure if run on default parameters.
A custom named data directory (in this example aucmedi.data) holds the two required
image directories: training for the training process and test for the prediction process.
aucmedi.data/
├── training/ # Required for training
│ ├── class_a/
│ │ ├── img_x.png
│ │ └── ...
│ ├── class_b/ # Subdirectory for each class
│ │ ├── img_y.png
│ │ └── ...
│ ├── class_c/
│ │ ├── img_z.png # Image names have to be unique
│ │ └── ... # between subdirectories
│ └── ...
├── test/ # Required for prediction
│ ├── unknown_img_n.png
│ └── ...
├── model/ # Will be created by training
├── xai/ # Will be created by prediction (optional)
├── evaluation/ # Will be created by evaluation
└── preds.csv # Will be created by prediction
The dataset structure is by default in the working directory for CLI or is mounted as volume into the container for Docker.
Dataset I/O Parameters¤
The dataset structure can be customized with the following parameters:
| AutoML Mode | I/O | Parameter | Default |
|---|---|---|---|
| Training | Input | path_imagedir |
'training' |
| Training | Output | path_modeldir |
'model' |
| Prediction | Input | path_imagedir |
'test' |
| Prediction | Input | path_modeldir |
'model' |
| Prediction | Output | path_pred |
'preds.csv' |
| Prediction | Output | xai_directory |
'xai' |
| Evaluation | Input | path_imagedir |
'training' |
| Evaluation | Input | path_pred |
'preds.csv' |
| Evaluation | Output | path_evaldir |
'evaluation' |
CSV Dataset (multi-label)¤
Instead of utilizing the directory interface for passing the training data class annotations via subdirectories, it is also possible to pass them as CSV.
This is required for setup a multi-label classification pipeline.
The default dataset structure then would look like this:
aucmedi.data/
├── training/ # Required for training
│ ├── img_x.png
│ ├── img_y.png # Single directory with all training images
│ ├── img_z.png
│ └── ...
├── test/ # Required for prediction
│ ├── unknown_img_a.png
│ ├── unknown_img_b.png
│ └── ...
└── annotations.csv # CSV annotations for training
The CLI command for multi-label CSV data would then look like this:
aucmedi training --path_gt annotations.csv --ohe
One-hot encoded CSV Example
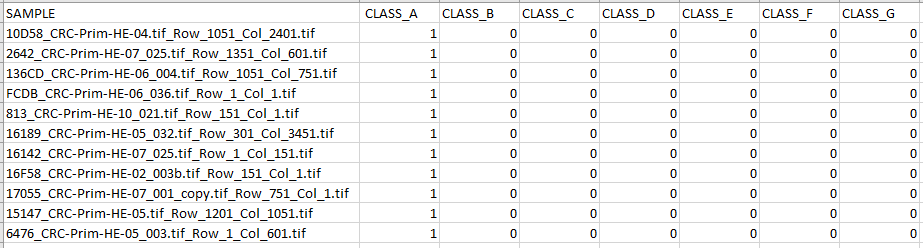
More information for CSV files can be found here: IO_CSV Interface