Quick Start - Framework
Pillar Structure¤
AUCMEDI is based on 3 pillars which allow building any state-of-the-art medical image classification pipeline.
Pillars of AUCMEDI
| Pillar | Type | Description |
|---|---|---|
| #1: input_interface() | Function | Obtaining general information from the dataset. |
| #2: NeuralNetwork | Class | Building the deep learning model. |
| #3: DataGenerator | Class | Powerful interface for loading any images/volumes into the model. |
The different pillars are represented in Python as function and classes. The passed parameters allow configuration of the automatically created image classification pipeline. The pillar interface of AUCMEDI combines intuitive and straightforward setup with the possibility of detailed custom specification.
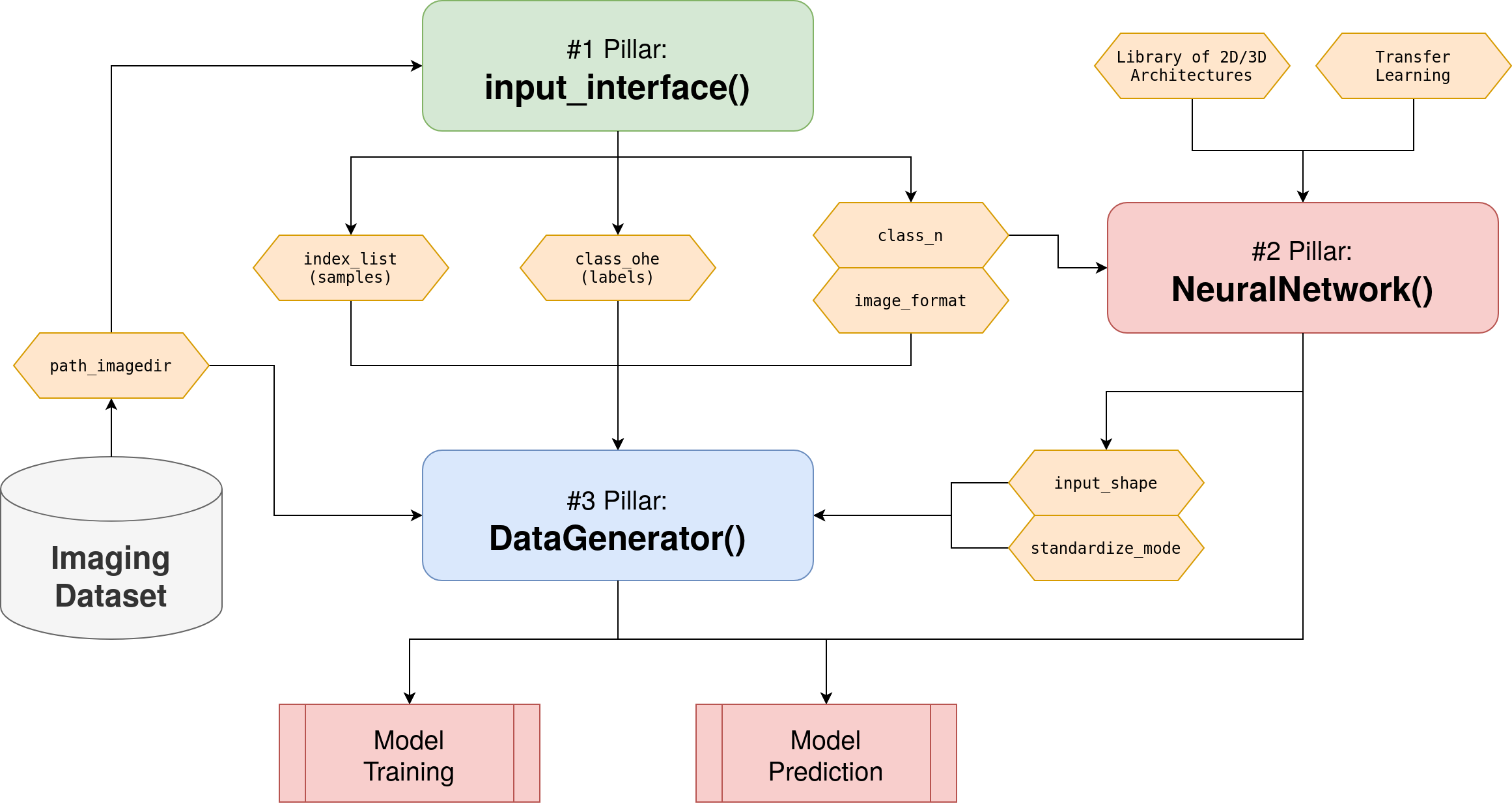 Flowchart diagram of the AUCMEDI pipeline showing the three pillars acting as access points: input_interface() for extracting relevant dataset information, NeuralNetwork for building the deep learning model and DataGenerator for data loading as well as preprocessing.
Flowchart diagram of the AUCMEDI pipeline showing the three pillars acting as access points: input_interface() for extracting relevant dataset information, NeuralNetwork for building the deep learning model and DataGenerator for data loading as well as preprocessing.
Basic Usage¤
In this example, a standard AUCMEDI pipeline build is demonstrated.
Install AUCMEDI via PyPI
Simply install AUCMEDI with a single line of code via pip.
pip install aucmedi
Build a pipeline
Let's build a COVID-19 Detection AI on 2D CT slides!
# AUCMEDI library
from aucmedi import *
# Pillar #1: Initialize input data reader
ds = input_interface(interface="csv",
path_imagedir="/home/muellerdo/COVdataset/ct_slides/",
path_data="/home/muellerdo/COVdataset/classes.csv",
ohe=False, # OHE short for one-hot encoding
col_sample="ID", col_class="PCRpositive")
(index_list, class_ohe, nclasses, class_names, image_format) = ds
# Pillar #2: Initialize a DenseNet121 model with ImageNet weights
model = NeuralNetwork(n_labels=nclasses, channels=3,
architecture="2D.DenseNet121",
pretrained_weights=True)
Train a model and use it!
Now, we want to use our pipeline to train it on our training data. Afterwards, we use our fitted model to classify 500 unknown ct slides.
# Pillar #3: Initialize training Data Generator for first 1000 samples
train_gen = DataGenerator(samples=index_list[:1000],
path_imagedir="/home/muellerdo/COVdataset/ct_slides/",
labels=class_ohe[:1000],
image_format=image_format,
resize=model.meta_input,
standardize_mode=model.meta_standardize)
# Run model training with Transfer Learning
model.train(train_gen, epochs=20, transfer_learning=True)
# Pillar #3: Initialize testing Data Generator for 500 samples
test_gen = DataGenerator(samples=index_list[1000:1500],
path_imagedir="/home/muellerdo/COVdataset/ct_slides/",
labels=None,
image_format=image_format,
resize=model.meta_input,
standardize_mode=model.meta_standardize)
# Run model inference for unknown samples
preds = model.predict(test_gen)
# preds <-> NumPy array with shape (500,2)
# -> 500 predictions with softmax probabilities for our 2 classes
More Details¤
More examples can be found here: Examples - Framework
The full documentation for AUCMEDI AutoML can be found here: API Reference